International Journal of Aquaculture and Fishery Sciences
Longterm Monitoring of Nitrification and Nitrifying Communities during Biofilter Activation of Two Marine Recirculation Aquaculture Systems (RAS)
Sabine Keuter1, Stefanie Beth2, Gerrit Quantz4, Carsten Schulz3,5 and Eva Spieck1*
2Landesbetrieb für Küstenschutz, Nationalpark und Meeresschutz Schleswig-Holstein, Herzog-Adolf-Str. 1, D-25813 Husum, Germany
3Gesellschaft für Marine Aquakultur mbH, Hafentörn 3, D-25761 Büsum, Germany
4Futurefish Aquaculture GmbH, Hansastr. 11, D-24118 Kiel, Germany
5Christian-Albrechts-University of Kiel, Institute of Animal Breeding and Husbandry, Hermann-Rodewald-Str. 6, D-24118 Kiel, Germany
Cite this as
Keuter S, Beth S, Quantz G, Schulz C, Spieck E (2017) Longterm Monitoring of Nitrification and Nitrifying Communities during Biofilter Activation of Two Marine Recirculation Aquaculture Systems (RAS). Int J Aquac Fish Sci 3(3): 051-061. DOI: 10.17352/2455-8400.000029Biofilters are crucial and costly components in marine recirculating aquaculture systems. However, not much is known about the settlement of nitrifying organisms and developing nitrification rates during the start-up phases of these reactors. The nitrifying microorganisms in moving bed biofilters of two marine recirculation aquaculture systems identical in construction were monitored for 388 and 477 days by PCR based methods, accompanied by laboratory nitrifying activity tests. Ammonia and nitrite were added to the recirculating aquaculture system 1, while system 2 was spiked with fish feed. On day 88, system 1 was stocked with turbot (at 17 °C) and on day 126 system 2 was stocked with sea bass cultivated at 22 °C. The potential nitrification rates corresponded well to the conditions in the operating systems, and in both systems slowly developing nitrite oxidation rates led to high nitrite peaks. However, after 218 (biofilter 1) and 286 (biofilter 2) days, potential rates of nitrite oxidizing bacteria outreached those of ammonia oxidizing bacteria.
The nitrite oxidizing bacteria were remarkably diverse on the genus level, and for the first time Nitrotoga was detected in marine biofilter systems. Nitrospira was assumed to be the most dominant nitrite oxidizing bacterium, also confirmed by electron microscopy. The ammonia oxidizing organisms belong almost exclusively to Nitrosomonas, of which dominant species shifted in both systems over time. The high similarities of some 16S rRNA gene sequences of Nitrospira and Nitrosomonas to sequences found previously in other marine recirculating aquaculture systems suggested that the species are characteristic for this artificial ecosystem.
Introduction
In recirculation aquaculture systems (RAS) only up to 10 % of the water is exchanged daily, hence toxic ammonia, the most relevant catabolic end product in intensive fish and shrimps production, has to be eliminated within the system to prevent fish toxic accumulation. The utilization of aerobic nitrification in moving bed filters of RAS was described first in the late 1980´s as summarized by Rusten et al. [1]. Despite its importance as system component as well as its costs, in-depth knowledge of such biofilters with respect to start-phases and malfunctions is still lacking [2]. Eshchar et al. [3] estimated that a nitrifying biofilter amounts for 20 % of the investment costs of a RAS, or $ 0.72 per kg fish produced per year. Therefore, Martins et al. [4] and Gutierrez-Wing and Malone [5] stressed the importance of further research to foster a development towards more efficient waste management which enables more economical and environmentally sustainable fish production in RAS.
Nitrification, the oxidation of ammonia to nitrate via nitrite, is performed by three phylogenetically distinct groups of chemolithoautotrophic microorganisms, the ammonia and nitrite oxidizing bacteria (AOB and NOB) and the ammonia oxidizing archaea of the novel phylum Thaumarchaeota (AOA) [6]. However, just recently NOB capable of ammonia oxidation have been identified [7,8], and sequences of these comammox genes have been found in freshwater RAS [9].
In the traditional taxonomy of nitrifiers, ammonia oxidizing microorganisms consist mainly of bacteria belonging to a monophyletic group within the Betaproteobacteria (Nitrosomonas, Nitrosospira), an exception are the exclusively marine representatives of Nitrosococcus belonging to the Gammaproteobacteria [10] and ammonia oxidizing archaea of the Thaumarchaeota. The guild of NOB is phylogenetically heterogeneous and so far comprises bacteria of six different genera: Nitrobacter (Alphaproteobacteria), Nitrospina (Nitrospinae), Nitrococcus (Gammaproteobacteria), Nitrospira (Nitrospirae), and the recently discovered Nitrotoga (Betaproteobacteria) as well as Nitrolancea (Chloroflexi) [11-13]. Both, AOB and NOB, are known to be fastidious and their cultivation is hampered by very slow growth rates. Therefore, the establishment of the nitrifying biofilm is tedious and hence involves economic losses in the start phases of RAS and elevated discharge of nitrogen-rich wastewater to the environment due to higher water exchange rates [1,14,15]. In comparison to each other, NOB have generally longer generation times, and are considered to be the weaker part of the nitrification process [16], and more sensitive towards deviating values of pH, oxygen concentrations, free ammonia concentrations etc. [17]. Hence, it is of special interest to study nitrifying performances of AOB and NOB separately, an approach so far not common in biofilter studies [18]. Therefore we applied activity tests with biocarriers (BC) in the laboratory for AOB and NOB respectively. These data were compared with in situ water quality parameters in the RAS. Species of nitrifying organisms that had settled on the biocarriers were characterized by 16S rRNA analyses throughout the time of the start-up phases. Besides bacterial nitrifies, also anaerobic ammonia-oxidizing planctomycetes (anammox) as well as ammonia-oxidizing archaea were sought after, since both have been recovered in moving bed biofilters of aquaculture systems [19,20].
The goal of this study was to characterize the critical start-up phases of nitrification biofilters of marine recirculating aquaculture systems (RAS), to correlate nitrifying efficiency of the biofilters with the presence of single nitrifying species and to investigate which feeding strategy (ammonia and nitrite versus fish feed) for the biofilter start-up is more reliable for nitrifying performance.
Materials and Methods
Description of the biofilters
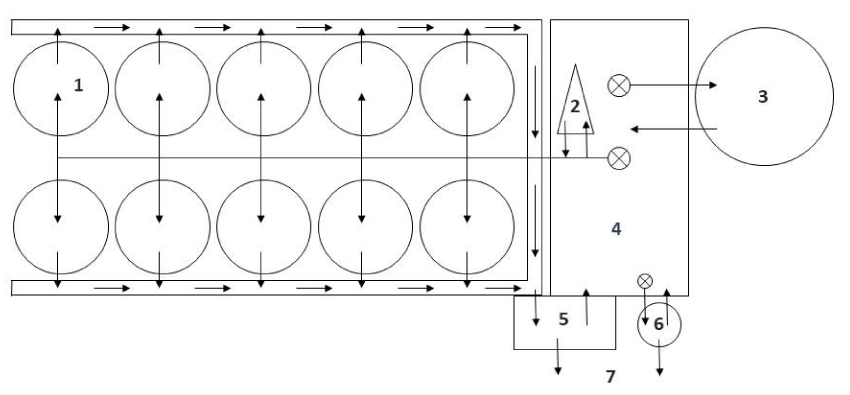
The RAS in this study are part of the marine aquaculture research facility of the GMA (Gesellschaft für marine Aquakultur mbH) located in Büsum at the North Sea coast of Schleswig-Holstein (Germany). Figure 1 shows a schematic diagram of the components of the modules. The total volume of the RAS modules is 35 m3 (25 m3 in fish tanks), the biofilters (BF) have a volume of 5.65 m3 each and were filled at the start with 1 m3 HDPE (high density polyethylene) biocarriers of the type 2H-BCN 012 KLL (GEA 2H Water Technologies, Wettringen, Germany) with a surface of 19.1 cm2 per biocarrier and 859 m2/ m3 and a protected (inner) surface of 704 m2/m3. The biocarriers were moved continuously by aerators arranged circularly at the bottom of the biofilter tank with an air flow of 98 m3 per hour. Before use, the biocarriers had been washed in freshwater (10 days), disinfected with a chloride-solution for a week and washed 2 to 3 times with freshwater. Additional 0.5 m3 of new biocarriers were added to the biofilter of RAS 1 on day 226, and to the biofilter of RAS 2 on day 302 and 351 to provide more area for the settlement of nitrifiers with the growing biomass in the tanks. The hydraulic retention times in the bio filters and the fish tanks were 8 and 24 min, respectively.
Further compartments of the RAS modules were drum filters (60 µm mesh size) and protein skimmers using ozone (Erwin Sander Elektroapparatebau GmbH, Uetze-Eltze, Germany). The latter were started only shortly before fish were stocked. The protein skimmers and the biofilters are individually connected to the pump sumps. From here, partly oxygenized water is pumped into the rearing tanks. Before returning to the pump sumps, the water is filtered by the drum filters (Figure 1). About 3 m3 system water is replaced daily with water from the adjacent port of Büsum, which is filtered and ozonated.
Addition of nitrogen and stocking with fish
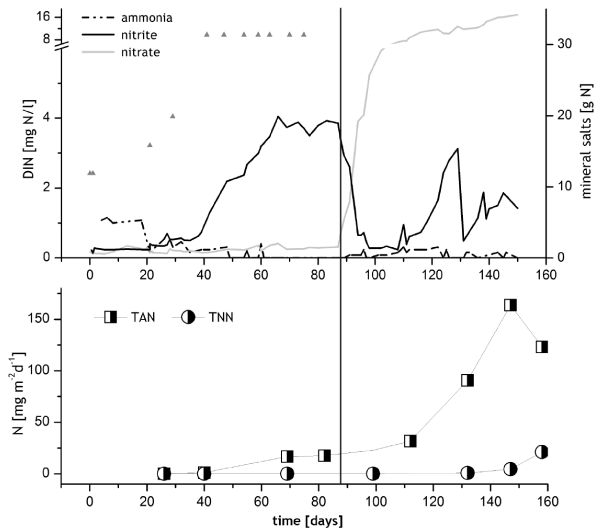
RAS 1: This module was filled with water on 02. July 2009. 30 g NH4Cl and 20 g NaNO2 (equaling 12 g N) were added on the first and second day to the pump sump, equaling 15% of the calculated 80 g TAN from the feeding rate of 100 kg turbot, which were originally expected to be introduced. When no ammonia was measured anymore, further portions of NH4Cl were given until day 75 (Figure 2) to maintain growth of biofilm. 21.3 kg (4,000) turbots (Psetta maximus) were stocked on day 88. With fish feed containing 54 % protein, approximately 15 mg TAN per m2 per day nitrification rate should have been reached by this time. Further 22.5 kg (3,000) fish were added on day 118 (43.86 kg in total). Although phosphate was not monitored, it was added as KH2PO4 (1.7 g) on days 0, 1, 41 and 47 to the pump sump. Since only nitrogen was added to RAS 1, a limitation of growth of nitrifiers by phosphorus was prevented. Another possible limiting factor for the growth of nitrifying bacteria in marine aquaculture systems is the trace metal iron, which is primarily bound to organic matter or precipitate as insoluble Fe (III) oxide-hydroxide in oxygenated seawater [21]. Since nitrifying bacteria are adapted to the use of this element despite its low concentrations in the open ocean, iron was not supplemented.
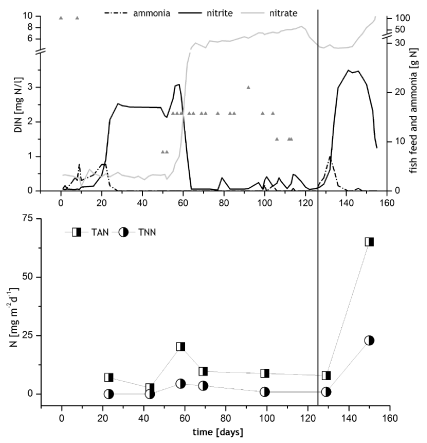
RAS 2: The second module was filled with water on 29. September 2010. 1.3 kg of fish feed pellets containing 45 % protein (equaling 101 g N) were added to the pump sump on day 1 and 6. Between day 50 and 113, 900 g NH4Cl were added in portions of up to 60 g (Figure 3). No phosphate was added here, since it is contained in fish feed (P = 1.7%). 5,000 (15 kg) sea bass (Dicentrarchus labrax) were stocked on day 126 and given feed with 48 % protein. With the initial feeding rate of 0.5 kg per day, the nitrification rate in biofilter of BF2 should have reached 25 mg TAN per m2 and day by stocking time.
h5>Water samples
Water samples were taken directly from the biofilters, first every 24h (from July 2009 until February 2010; except weekends), then every 48h. Sampling periods were 477 days (RAS 1) and 388 days (RAS 2) resulting in 282 and 208 samples (as duplicates) for RAS 1 and RAS 2, respectively. Ammonia was measured with the powder-pillow tests and the DR 2800 spectrophotometer (both Hatch Lange, Düsseldorf, Germany). For nitrite and nitrate concentration measurements by ion pair chromatography (Elite LaChrom System, Hitachi, Krefeld, Germany), the samples were diluted 1:5 with water to avoid interfering chloride peaks [22]. Measurements of the following water quality parameters were included in this study: oxygen and temperature (Oxyguard Polaris, Birkerød, Denmark), redox potential (Aquamedic mV Controller, Bissendorf, Germany), pH (1200-S Sensor, Hach Lange, Düsseldorf, Germany).
Nitrogen input
The daily average nitrogen input in form of fish feed for each month was calculated using the equation of Timmons et al. [23], PTAN = F * PC * 0.092 with PTAN = total ammonia nitrogen production rate (kg per day), F = feed rate (kg/day) and PC = protein content in feed (decimal value).
Activity tests
Sampling for activity tests and DNA extraction started around day 20 of each biofilter. In the first half year, samples were taken almost fortnightly, later every 4th to 8th week, in total 17 (until day 477) from BF 1 and 12 times from BF 2 (until day 388), respectively. The activity tests were conducted following the method of Spieck and Lipski [22]: in parallels, 5 biocarriers were shaken (at 170 rpm) in 25 ml marine mineral salts medium (AOB medium: 5.4 mg KH2PO4; 1 mg FeSO4 x 7 H2O; 1 ml NOB trace elements solution; 5 g CaCO3; 700 ml seawater; 300 ml aqua dest; NOB medium was prepared according to Watson and Waterbury [24]). The media were spiked with 0.5 or 1 mM ammonia (AOB) or nitrite (NOB) as substrates. Nitrite and nitrate concentrations were measured with HPLC. Nitrate production served as substrate conversion of NOB and the sum of nitrite and nitrate as substrate conversion of AOB, since the stoichiometrical conversion of ammonia over nitrite to nitrate is 1:1:1. The maximum nitrification potentials per area were calculated for AOB and NOB respectively, using the average of two parallels and the total area of biocarriers, not the so called protected area. These are biocarrier-specific rates. Biofilter-specific nitrification rates were calculated by converting area-specific rates of the biocarriers into the total biocarrier surface area in the biofilter. The calculations were adjusted according to the further additions of biocarriers.
DNA extraction
Of each sample, taken together with the samples for the activity tests, 10 biocarriers were counted in 50 ml plastic tubes with 10 ml 2 % NaCl, 0.1 % Triton X 100 and 2.5 g of each 1.7-2 mm and 0.45-0.5 mm glass beads. After sonication (1 h) the tubes were shaken vigorously for 1 h before the liquid phase was centrifuged. DNA was extracted from the pellets using the MoBio soil DNA extraction kit (MoBio Laboratories, Carlsbad, CA). For some samples the MoBio biofilm DNA extraction kit and the QIAmp DNA stool mini kit (Quiagen, Hilden, Germany) were used additionally to check whether these kits might extract DNA of more or different nitrifying bacteria. Sample names compose of the biofilter number (BF1 or BF2) and the time of the sample taken, i.e. BF1-99 is a sample of biofilter 1 taken on day 99 after start.
PCR, DGGE, cloning
Extracted DNA was screened using group specific primer pairs targeting 16S rRNA gene sequences of nitrifying bacteria and amoA gene sequences of archaea (Table 1). PCR were conducted with purified 16S rRNA amplicons from previous PCR (nested PCR) except for PCR targeting the gene sequences of enzymes. Where possible, PCR products were directly sequenced after purification. The Nitrospira-specific PCR-product (27f/1158r) of sample BF2-344 was cloned and 40 clones were screened using RFLP with the restriction enzyme HpaII.
For denaturing gradient gel electrophoresis (DGGE) [25], 16S rRNA gene fragments were amplified with the GC-clamped primer pairs CTO189f/CTO654r (ß-AOB, [26]). The products were loaded into a gel of 40–70% denaturants, and run at 110 V and 59 °C for 17 h. Bands of interest were excised, re-amplified and sequenced. The sequences were analyzed using BLASTN [27] and the MEGA software [28]. Sequences mentioned below were deposited in GenBank under the accession numbers JN903955-JN903972.
The primer pair Nspina190/934 (forward: 5´-CTCAAAGATGAGTTCGCGGACC-3´; reverse: 5´- CCCAAGCAAGTCAAATCCAG -3´; E. coli positions 78–470; annealing temperature 68°C) was deduced using the program Clone Manager (Sci-Ed Software, Cary, USA) and evaluated with the Basic Local Alignment Search Tool (BLAST, NCBI) and Probe Match [29].
Electron microscopy
Pellets of cells (see DNA extraction) of BF1-477 and BF2-388 were fixed in 2.5% (v/v) glutaraldehyde for 1.5 hours and 2 % (w/v) osmium tetroxide overnight, and embedded in Spurr [30]. Ultrathin sections were stained with uranyl acetate [31] and lead citrate [32] and inspected by transmission electron microscopic (Zeiss model Leo 906E, Carl Zeiss, Jena, Germany).
Results and Discussion
Nitrifying microbial community analysis
The microbial nitrifying communities attached on the filter media were analysed using PCR based approaches supported by electron micrographs of ultrathin sections of cells from the bio carriers. Most PCR amplicons could be sequenced directly after group-specific PCR, indicating rather dominant representative of the respective genera or species (Table 1).
The 16S rRNA gene analyses revealed that representatives of most of the known genera of autotrophic nitrifying bacteria were present in the biofilters examined. Only for the Gammaproteobacteria Nitrococcus and Nitrosococcus PCR results were negative or unclear (amplification of correct sequence size but poor sequences). Nitrospina-specific PCR runs were positive for several samples, however, it was only possible to retrieve good sequences from sample BF1-477 (= sample of day 477; 444 bp) and BF2-388 (= sample of day 388; 263 bp), which were 95 % similar to a clone from a Nitrospina enrichment culture (KC706458; North Sea water). It is not clear, why only few good sequences could be obtained from Nitropina-specific PCR, it is possible that the relatively high annealing temperatures produced secondary priming artefacts and noisy sequences. The information about Nitrospina species in aquaculture and in general is very valuable, therefore more appropriate primers should be designed for future studies. Primers targeting 16S rRNA gene sequences of planctomycetes led to amplicons of the right lengths in several samples from both filters. Two (BF1-477 and BF2-99) were sequenced, but only BF1-477 was 85.6 % (265 bp) similar to Cand. Brocadia fulgida, a bacterium known to perform anammox.
Ammonia-oxidizing bacteria
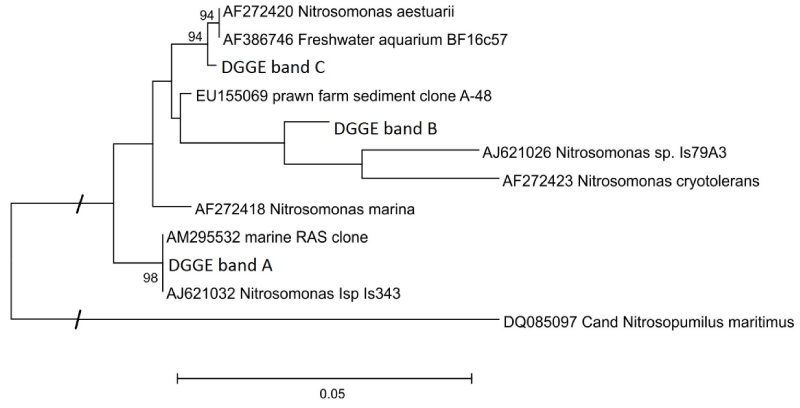
From sampling days 54 (BF1) and 23 (BF2) on sequences of AOB belonging to the subclass Betaproteobacteria (ß-AOB) were obtained by DGGE. The AOB populations in both bio- filters changed, though at different times and differently quick (Figure 4). In total, three dominant bands (A-C) could be detected during the sampling period: band A and B in BF1 and B and C in BF2. Interestingly, the dominant band of BF1 changed from A to B between February (day 239) and April (day 279). Until February, only band B was detected in BF2 (day 129), then band C appeared and both populations coexisted until May, when the first population (band B) disappeared. Further faint bands that could be observed occasionally were heterodupleces and of the same sequences as the adjacent dominant bands.
Known key factors influencing AOB population structures are temperature [33], ammonia concentrations [34], salinity [35, 36] or oxygen [37]. At the time of stocking in RAS 2, the temperature has been lowered enormously, salinity dropped and more ammonia became available with the fish feed, factors which could have triggered the population shift. A further possibility is that the fish might have transferred a highly competitive AOB from the hatchery system.
In BF1 the AOB population shift concurred with a strong decrease of AOB activities (Figure 4), which occurred due to a feeding stoppage (for the tagging and sorting of the fish). The shift in AOB population was linked previously to changes in ammonia removal rates in WWTP [38] as well as in marine gravel biofilters [39].
The sequences recovered from bands of several DGGE-gels (approx. 400 bp) were identical, respectively, corresponding to their position in the gel. PCR products amplified with the primer pair Nso190/NitB were sequenced directly, and since these sequences were identical with the corresponding bands from the DGGE approach, longer sequences of approx. 1000 bp could be phylogenetically analysed (Figure 5). All three sequences can be allocated to the Nitrosomonas cluster 6 with similarities of 97.1 % (A), 95.7 % (B) and 97.8 % (C) to N. marina (accession number: AF272418). More detailed, the sequence of band A is identical to a sequence derived from a marine RAS in Israel (AM295532), and 99.8 % similar to the strain Is343 (AJ621032); the sequence of band B is most similar (96.6 %) to a clone derived from prawn farm sediments (EU155069), the closest described relative is the Nitrosomonas strain Is79A3 (96.1 %; AJ621026); the sequence of band C is similar to the isolate BF16c57 from a freshwater aquarium (AF386746) and to N. aestuarii (AF272420) with 99.4 % each (Figure 5). In comparison with the 805 bp long sequence of a clone of a bacterium enriched in a fluidized bed reactor inoculated with water from the port of Büsum (FN394309, [40]) the corresponding sequence fragment of band C was almost identical (99.9 %). ß-AOB specific DGGE with DNA extracted with biofilm or stool kit did not result in further or different bands. Direct PCR with Nitrosospira-specific primers led to positive signals with several samples between day 22 and 188 from both biofilters; however, only from sample BF1-188 on an acceptable sequence of 405 bp could be retrieved, that was similar to a Nitrosospira-like sequence (99.7 %; Nitrosospira sp. Nsp17; AY123804).
The diversity of ammonia oxidizers´ populations in WWTP varies among studies between single populations to the coexistence of several different AOB and AOA [41 and therein]. The occurrence of Nitrosospira sp Nsp17 sequences in BF1 could indicate that the size of the Nitrosospira population is below the detection limit of DGGE (less than 1 % of total cells) [23]. Nitrosospira-like bacteria have been found as dominant AOB in biofilters [42-44] or as only AOB in a marine RAS [45], although they are rather characterized as dominant AOB in habitats with low substrate concentrations [46] and other adverse conditions like low DO, short HRT, allythiourea [47]. AOB as well as AOA were found together in marine aquaria and RAS [33,45,48,49]. However, no Archaea at all were found in the biofilters with primers targeting archaeal 16S rRNA as well as archaeal amoA (encoding subunit A of the ammonia monooxygenase).
Nitrite oxidizing bacteria
The NOB species in the two biofilters were highly diverse on the genus level, and more than in any other aquaculture study so far, including next generation sequencing approaches [49,50].
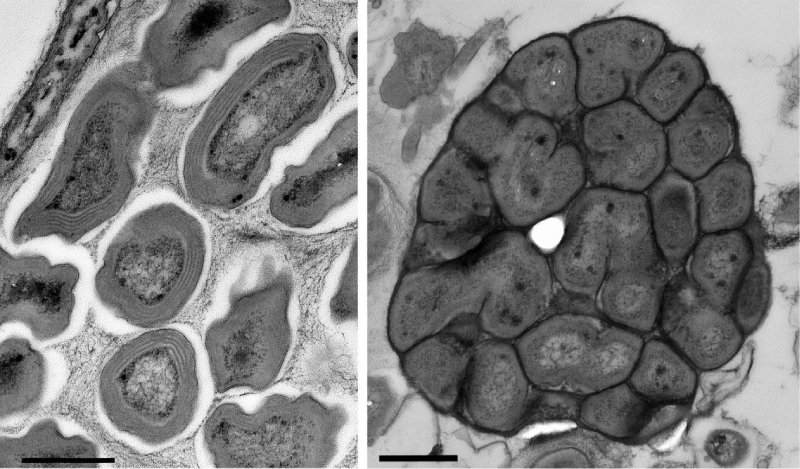
Nitrospira was identified as the dominant NOB in marine RAS [48,51-53] and this is also indicated by the results of this study. Samples were not quantified in detail in this respect. However, by electron microscopic inspection of the samples BF1-477 and BF2-365 only Nitrosomonas- and Nitrospira-like cells were found in colonies throughout the ultrathin sections, although Nitrobacter and Nitrotoga would have been easy to identify (Figure 6). Furthermore, all Nitrospira-specific PCR runs were positive from the first to the last sampling days, which was not the case for Nitrobacter and Nitrotoga.
Nitrospira positive PCR products of early samples could not be sequenced directly (mixed sequences) except for BF1-40 (533 bp; 97.0 % similar to N. moscoviensis). Sequences (between 900 and 1160 bp) from later samples of the biocarriers from both biofilters (BF1-478, BF2-344) were 99.3 to 100 % identical to the strain Ecomares 2.1 (HQ686082; [52]), a close relative of Nitrospira marina isolated from another marine RAS in Büsum. High similarities (98.9-99.7 %) were also found when the sequences were compared to the 713 bp long sequences of clones of bacteria from the before mentioned saline fluidized bed reactor (FN394312-14; [40]).
The very specific primer pair “M1 mar” targets the sequence of a clone retrieved from the mentioned RAS in Büsum [53]. It matched only with the DNA of samples of BF2 on day 344 and 399, and the sequence 344 (313 bp) is 98 % similar to the sequence of the clone “M1 mar” and sequences from a RAS at the Baltic Sea [54]. With DNA of the same sample, clones were produced using the semi-specific Nitrospira-primer pair 27f/1158r. 40 of these clones were screened by RFLP and all were of the same sequence, similar to the Ecomares 2.1 strain (99.3 %; 1150 bp).
Generally, Nitrospira sequences found in the biofilters´ biofilm samples were similar to sequences found in before mentioned marine or brackish RAS including the non-marine Nitrospira-like sequence (N. moscoviensis) [48,52-54], although in a 454 pyrosequencing approach N. moscoviensis sequences were by far more abundant than those of the other Nitrospira strains [50]. The results of the RFLP confirmed a dominant role of the strain Ecomares 2.1 over other Nitrospira in the later biofilm samples of BF2. These findings corroborate the suggestion that the strain is characteristic for its habitat [53].
PCR runs with Nitrobacter-specific primer pairs were positive with samples from both biofilters from day 20 on for the whole sampling period (with 2 and 3 exceptions in summer). Amplicons from several samples were sequenced resulting in three slightly different, 245 bp long rRNA gene sequences (98-99.5% similarity to each other). The sequence retrieved from the samples BF1-26, BF1-279, and BF2-23 was 99.1% similar to the marine Nitrobacter strain 311 (AM292300) and 99.6% to the Nitrobacter sp strain PBAB17 (AY508476); the sequence of BF1-188 and BF2-99 was identical with a Nitrobacter enriched from a wastewater treatment plant (FJ263062, [55]) and the sequence of the samples BF1-478, and BF2-388 was identical with N. vulgaris (AM292301) and N. hamburgensis (CP000319). Concerning the putative diversity within the genus Nitrobacter, it is problematic to determine the species by comparing 16S rRNA gene sequences, since these do not differ much between the phylogenetically young Nitrobacter [56]. The differences described before comprise of about 4 nucleotides within the relatively short sequences.
The specific primers targeting Nitrotoga-like bacteria amplified sequences of the expected size with DNA from sample BF1-26 and BF2-58 on almost constantly. A total of 7 sequences (~575 bp) was recovered from samples between days 58 and 388 and all were identical to each other and almost identical to Cand. Nitrotoga arctica (99.7 %; DQ839562).
The existence of the nitrite oxidizing betaproteobacterium Nitrotoga was discovered only in 2007 [57]. Following reports of Nitrotoga in biofilters or reactors linked its appearance with cold temperatures [57-59], but reports with findings of Nitrotoga related sequences in marine habitats are very scarce. Only recently was a non-marine Nitrotoga enriched from eelgrass sediment growing best at 16°C, with a 16S rRNA gene similarity of 99.6 % to Cand. Nitrotoga arctica and 99.3 % to the sequences in this study [60]. Thus, the frequent detection of this nitrite oxidizer especially in the relatively warm biofilter of RAS 2 was quite surprising and should encourage future research to consider the use of Nitrotoga-specific primers and probes when analyzing nitrifying communities in marine environments. To investigate the origin of Nitrotoga, 150 ml of marine NOB medium were inoculated with 50 ml of the filtered and ozonated sea water used for the RAS modules. Obtained sequences of the growing NOB enrichment verified that the Nitrotoga stems from the North Sea water in the port of Büsum and did not enter the bio filters via water/fish additions (data not shown). It seems conceivable that the boundary between marine and non-marine lifestyle of Nitrotoga is less strict than thought, which is possibly combined with certain circumstances and requires further studies with cultures obtained from the RAS.
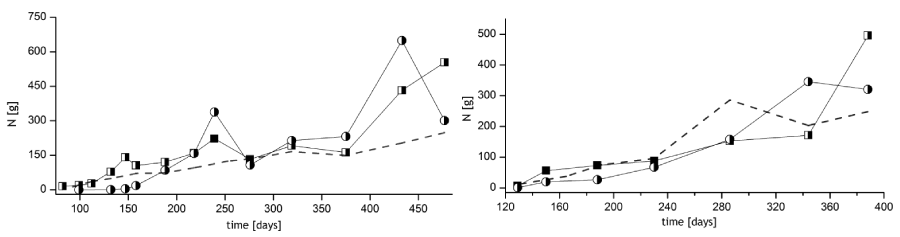
Mainly due to the different fish species, the modules exhibited a range of differences after stocking (Table 2). However, the compositions of the nitrite oxidizing bacterial populations, in contrast to the AOB, were very similar and did not change considerable over time. Wittebolle and colleagues [41] assumed that the inoculum (in this case seawater from the port of Büsum) is more important for the further composition of nitrifying bacteria than the operational parameters of biofilters. This is in accordance with the very high similarities with sequences of nitrifying bacteria found by Sudarno and his colleagues [40] in the saline FBR filled with North Sea water.
Potential nitrification activity tests
The ammonia and the nitrite oxidation potentials of biocarriers were obtained in laboratory tests and are shown in the lower graphs in figures 2 and 3 as the oxidized TAN (total ammonia nitrogen) and TNN (total nitrite nitrogen) per m2 and day (biocarrier-specific rates). The two parallels in these tests were always almost identical, thus standard deviations are omitted. In all tests the concentration of the end products equaled the concentration of the substrates given, thus no fractions of other DIN species were produced by different reactions than nitrification. The potential rates could be correlated with the monthly average feed input per day, which indicates that the activity tests corresponded to the in situ nitrification rates in the operating system. The correlation coefficients between the nitrogen input into the system and the development of AOB and NOB potential activities were in the range of 0.89 and 0.99 before stocking and 0.78 and 0.95 after stocking (Table 2). The potential nitrification tests also reflected temporary feeding interruptions, as observed e.g. for RAS 1 on day 276, were the activities of AOB as well as NOB were substantially lower compared to the previous sampling day (Figure 4). Before day 276 the fish were not fed for almost two weeks.
Development of nitrification in the biofilters
In BF1 1 mg TAN/l was measured until day 20, then the ammonia peak decreased and nitrite concentration increased. From day 59 until day 88, a nitrite peak of between 3 and 4 mg/l TNN persisted (Figure 2). On day 87, one day before the first stocking, the foam fractioner and thereby ozone addition was started, the nitrite concentration decreased and the nitrate concentration started to increase. After day 118, when further turbots were stocked, another nitrite peak developed which was less pronounced and decreased after fewer days. No ammonia peak evolved after stocking. The average temperature was 22.3 °C until the fish arrived and 17.2° C after stocking (Table 2), and was temporarily reduced to a minimum of 14.4 °C (day 102) for the acclimatization of the animals. Nitrifying potentials of AOB on the biocarriers from module RAS 1 were measured on day 40 with 1.14 mg TAN per m2 *d-1, and when the first batch of turbot (21.3 kg) was brought in, potential ammonia oxidizing activities of the biocarriers were 17.6 mg TAN per m2 *d-1 (day 82), matching the calculated aim of 15 mg TAN per m2 *d-1. NOB activities on the BC in BF1 developed only several weeks after the stocking but then increased from 21 to 99 mg TNN per m2 *d-1 between day 158 and 188 and overtook the AOB on day 218 with 184 mg TNN m2 *d-1 (Figure 4).
In RAS 2, concentrations of >0.3 mg/l TAN were measured between day 6 and 23 (Figure 3). With the subsequent decrease of ammonia concentration, caused by early AOB activity, a nitrite peak (2-3 mg/l TNN) developed and persisted for almost 40 days. The peak then decreased quite rapidly, simultaneously with the onset of some NOB activity (4.4 mg TNN per m2 *d-1 on day 58) and the increase of nitrate concentrations (Figure 3). For the acclimation of the fish the temperature was reduced from 24.2 to 12.7 °C between day 111 and 127, the salinity dropped from 37 to 29 ppt in the same time (not shown). On day 154, the temperature reached again 20°C and salinity was 30 ppt. The foam fractioner and ozone addition were started on day 105.
The nitrification potentials of the AOB increased earlier and faster, most probably due to the faster growth rates. Satoh et al. [18] reported a delayed growth of nitrite oxidizers compared to AOB despite of the initial presence of nitrite and their rate measurements showed further, that the addition of nitrite had no advantage for NOB growth compared to the control without nitrite. This is in accordance with the results from RAS 1, which was also spiked initially with nitrite.
Comparisons of nitrifying activities in the pre-fish phase of the biofilters indicated an advantage of using fish feed as inoculum to facilitate the growth of nitrifying bacteria. In general, a strong increase of organic matter can shift the C/N ratio, which favors the proliferation of heterotrophic bacteria in comparison to nitrifiers [61]. Ammonia and nitrite oxidizing bacteria are chemolithoautotrophic microorganisms, which have the genetic inventory to assimilate simple organic substances like pyruvate [62, 11], but are inhibited by a wide range of organic molecules [63]. Therefore, dissolved degradation products of the fish feed can have a negative impact on the nitrifying performance. Further on, especially in marine systems, high levels of sulfate provide the basis for the production of sulfide [64] which is a well-known inhibitor of nitrification revealing a greater effect on nitrite oxidation than on ammonia oxidation [65]. As a consequence, over-feeding should be avoided to stabilize nitrification.
However, the idea of using fish feed was that the nitrifying biofilm could be already adapted to a certain extent to the condition in the system after fish introduction, e.g. in terms of organic substances. This consideration was taken into account, since it was observed that nitrification potentials of biofilters break down for unknown reasons after the introduction of fish ([5], G. Quantz, personal communication). A further effect could be that EPS (extracellular polymeric substances) formed by heterotrophic bacteria are used by slowly growing nitrifying bacteria. Tsuneda et al. [66] investigated an enhancing effect on nitrifying biofilm formation with the help of EPS produced by heterotrophs. They assumed that the heterotrophs transform interfacial properties in favor of nitrifying bacteria.
After the systems were stocked with fish, ammonia oxidizing activities increased quickly in both biofilters and ammonia concentrations remained acceptable. In contrast, high nitrite peaks occurred in both modules, so that fish feed input could not be increased as planned and in RAS 2 water had to be exchanged massively between day 135 and 148 (data not shown). The variety of known and unknown parameters differing between the modules, mainly due to different fish species and therefore different treatments, leaves little room for speculations with regards to the inoculation method for the post-stocking phase.
Similar in both modules was the rapid decrease of water temperature for the acclimatization of the fish. This might have caused some stress to the young nitrifying biofilm and reduced growth in BF1 in the further run at temperatures around 17 °C. NOB activities as measured in the laboratory were very low at the time of stocking and the NOB could probably not keep up with the increasing nitrite production, due to their genuinely slow growth rates, which led to nitrite peaks after stocking.
Scrutinizing the course of nitrite concentrations, NOB activities and the onset of ozone addition in RAS 1, it becomes clear, that ozone has an inhibitive influence on the already slow growth of the NOB. Nitrite reacts with ozone to nitrate especially when further competitive substrates for the reaction with ozone (e.g. yellow substances) are absent [67], leading to a deprivation of substrate for bacterial nitrification [68]. In RAS 1, high nitrite peaks were replaced by steeply increasing nitrate concentrations just after ozone addition had started on day 87 (Figure 2), while NOB activities remained low. These circumstances indicate nitrite elimination by ozone, which competes for nitrite with the slow growing NOB. Nevertheless, with increasing concentrations of other substrates for ozone, the nitrite oxidation potentials increased remarkably after day 158.
Around day 218 (BF1) and 286 (BF2), the potential nitrite oxidation activities in the biofilters reached those of the ammonia oxidation activities and even exceeded them in the further course (Figure 4). Higher activities of NOB have been observed regularly in our lab, but also in other studies where the nitrification rate measurements have been decoupled [69]. Correspondingly, abundances of NOB cells have been reported to be higher than that of AOB in nitrifying communities. [52,70]. This might be explained by the high versatility of NOB, or specifically Nitrospira, regarding their substrates. Some Nitrospira species are capable of growing alternatively with hydrogen, formate or urea or oxidize ammonia [7,8,71]. Using other substrates than nitrite, and especially highly available ammonia, NOB would have an advantage over AOB to grow in higher abundances, which lead to higher rates in the activity tests. In a freshwater RAS, Nitrospira genes of comammox, the complete ammonium-to-nitrate oxidation, have been detected [9], but not in marine systems. This theory has yet to be scrutinized.
In general, the investigation of nitrification in biofilters is complex and has its limitations, both on bench-scale, with results that cannot be linked directly to systems, and on full-scale, with uncontrollable parameters and hardly interpretable results [73]. Our study attempts to link laboratory activity tests, processes in full scale biofilters and microbial diversity, and was likewise limited by dissimilar and uncontrollable parameters in the operating system stocked with animals and the restricted amount of set-ups.
Conclusion
The data obtained from the rare opportunity to longterm sample two identical production-scale marine RAS from the start gave new insights into the development and composition of the nitrifying microorganism community and the unsteady development of ammonia and nitrite oxidizing rates. We could show that the laboratory activity tests can be used to estimate the biofilter performance capacity and with the results underline the importance of AOB-NOB discriminating research, especially since the discovery of Nitrospira species capable of comammox. The nitrifying community was very diverse on NOB genus level including the detection of marine representatives of Nitrotoga. In general, knowledge of the microbial community in a man-made system that relies on the physiology of its members will be useful for operational and constructional improvements of marine RAS biofilters.
The authors would like to thank Elke Woelken, University of Hamburg, for excellent technical assistance in electron microscopy and Markus Griese and Michael Schlachter for sampling and water quality data collection. Thanks are further due to our project partners LimnoMar (Hamburg), GEA 2H Water Technologies (Wettringen), Spranger Kunststoffe (Plauen) and the working group “Aquatic Biotechnology” of Prof. Wolfgang Sand at the Biofilm Centre of the University of Duisburg-Essen. This work was funded by the Deutsche Bundesstiftung Umwelt (AZ 23821/01-03).
- Rusten BB, Eikebrokk B, Ulgenesb Y, Lygrenc E (2006) Design and operations of the Kaldnes moving bed biofilm reactors. Aquacult Engin 34: 322-331. Link: https://goo.gl/SrDJh
- Zohar Y, Tal Y, Schreier H, Steven C, Stubblefield J, et al. (2005) Commercially feasible urban recirculating aquaculture: addressing the marine sector. In Urban aquaculture. Edited by Costa-Pierce B, Desbonnet A, Edwards P, Baker D, CABI Publishing, Oxfordshire UK 159-171. Link: https://goo.gl/KMp48T
- Eshchar M, Lahav O, Mozes N, Peduel A, Ron B (2006) Intensive fish culture at high ammonium and low pH. Aquaculture 255: 301-313. Link: https://goo.gl/3mvDhb
- Martins CIM, Eding EH, Verdegem MCJ, Heinsbroek LTN, Schneider O, et al (2010) New developments in recirculating aquaculture systems in Europe: A perspective on environmental sustainability. Aquacult Engin 43: 83-93. Link: https://goo.gl/9nST9Q
- Gutierrez-Wing MT, Malone RF (2006) Biological filters in aquaculture: Trend and research directions for freshwater and marine applications. Aquacult Eng 34: 163 -171. Link: https://goo.gl/VBa4Qr
- Spang A, Hatzenpichler R, Brochier-Armanet C, Rattei T, Tischler P, et al. (2010) Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol 18: 331-40. Link: https://goo.gl/QbPXC2
- Daims H, Lebedeva EL, Pjevac P, Han P, Herbold C, et al. (2015) Complete nitrification by Nitrospira bacteria. Nature 528: 504–509. Link: https://goo.gl/VewUYJ
- Van Kessel MA, Speth DR, Albertsen M, Nielsen PH, Op den Camp HJM, et al. (2015) Complete nitrification by a single microorganism. Nature. Link: https://goo.gl/ouz2NV
- Bartelme RP, McLellan SL, Newton RJ (2017) Freshwater recirculating aquaculture system operations drive biofilter bacterial community shifts around a stable nitrifying consortium of ammonia-oxidizing archaea and comammox Nitrospira. Frontier Microbiol 8: 1-18. Link: https://goo.gl/bXhvgR
- Koops HP, Purkhold U (2006) The lithoautotrophic ammonia-oxidizing bacteria. In The Prokaryotes. Edited by Dworkin M, Falkow S, Rosenberg E, Schleifer K-H Stackebrandt E Springer New York. 778-811. Link: https://goo.gl/GNKEh6
- Starkenburg SR, Spieck E, Bottomley PJ (2011) Metabolism and genomics of nitrite oxidizing bacteria: Emphasis on pure culture studies and Nitrobacter species. In Nitrification. Edited by Ward BB, Arp DJ, Klotz MG, ASM Press, Washington, D.C: 325-345. Link: https://goo.gl/jiJ6Z1
- Sorokin DY, Lücker S, Vejmelkova D, Kostrikina NA, Kleerebezem R, et al.(2012) Nitrification expanded: discovery, physiology and genomics of a nitrite-oxidizing bacterium from the phylum Chloroflexi. Isme J 6: 2245-2256. Link: https://goo.gl/MJ1tFs
- Spieck E, Keuter S, Wenzel T, Bock E, Ludwig W (2014) Characterization of a new marine nitrite oxidizing bacterium, Nitrospina watsonii sp. nov, a member of the newly proposed phylum "Nitrospinae". Syst Appl Microbiol37: 170-176. Link: https://goo.gl/ffLJVH
- Nijhof M, Bovendeur J (1990) Fixed film nitrification characteristics in sea-water recirculation fish culture systems. Aquaculture 87: 133-143. Link: https://goo.gl/25MUft
- Gross A, Nemirovsky A, Zilbergb D, Khaimova A, Brennerc A, et al. (2003) Soil nitrifying enrichments as biofilter starters in intensive recirculating saline water aquaculture. Aquaculture 223: 51-62. Link: https://goo.gl/3yUarx
- Graham DW, Knapp CW, Van Vleck ES, Bloor K, Lane TB, et al. (2007) Experimental demonstration of chaotic instability in biological nitrification. Isme J 1: 385-393. Link: https://goo.gl/WFLefb
- Villaverde S, García-Encina PA, Fdz-Polanco F (1997) Influence of pH over nitrifying biofilm activity in submerged biofilters. Wat Res 31: 1180-1186. Link: https://goo.gl/6cLfhk
- Satoh H, Okabe S, Yamaguchi Y, Watanabe Y (2003) Evaluation of the impact of bioaugmentation and biostimulation by in situ hybridization and microelectrode. Wat Res 37: 2206-2216. Link: https://goo.gl/K5req8
- Tal Y, Watts JEM, Schreier SB, Sowers KR, Schreier HJ (2003) Characterization of the microbial community and nitrogen transformation processes associated with moving bed bioreactors in a closed recirculated mariculture system. Aquaculture 215: 187-202. Link: https://goo.gl/nbsfG4
- Van Kessel MA, Harhangi HR, Flik G, Jetten MS, Klaren PH, et al. (2011) Anammox bacteria in different compartments of recirculating aquaculture systems. Biochem Soc Trans 39: 1817-1821. Link: https://goo.gl/54mVN4
- Kuma K, Nishioka J, Matsunaga K (1996) Controls of iron (III) hydroxide solubility in seawater: The influence of pH and natural chelators. Limnol Oceanogr 41: 396-407. Link: https://goo.gl/zoEpNk
- Spieck E, Lipski A (2011) Cultivation, growth physiology, and chemotaxonomy of nitrite-oxidizing bacteria. In Methods in Enzymology 486: 109-130. Link: https://goo.gl/LY6JsY
- Timmons MB, Ebeling JM, Wheaton FW, Summerfelt ST, Vinci BJ (2002) Recirculating Aquaculture Systems. Cayuga Aqua Ventures, Ithaca, New York.
- Watson SW, Waterbury JB (1971) Characteristics of two marine nitrite oxidizing bacteria, Nitrospina gracilis nov. gen. nov. sp. and Nitrococcus mobilis nov. gen. nov. sp. Arch Microbiol 77: 203-230. Link: https://goo.gl/AmH4Jh
- Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695-700. Link: https://goo.gl/i6zDK9
- Kowalchuk G, Stephen J, De Boer W, Prosser J, Embley T, et al. (1997) Analysis of ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl Environ Microbiol 63: 1489-1497. Link: https://goo.gl/PzYqbp
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410. Link: https://goo.gl/NMNMM4
- Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596-1599. Link: https://goo.gl/JSV23M
- Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, et al. (2007) The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res 35: D169-D172. Link: https://goo.gl/SjGcTv
- Spurr AR (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastr Res26: 31-43. Link: https://goo.gl/BHtzJD
- Watson M L (1958) Staining of tissue sections for electron microscopy with heavy metals. J Biophys Biochem Cytol 4: 475-478. Link: https://goo.gl/tDmLtr
- Reynolds ES (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17: 208-212. Link: https://goo.gl/n7WRBr
- Urakawa H, Tajima Y, Numata Y, Tsuneda S (2008) Low temperature decreases the phylogenetic diversity of ammonia-oxidizing archaea and bacteria in aquarium biofiltration systems. Appl Environ Microbiol 74: 894-900. Link: https://goo.gl/XZbsx4
- Lydmark P, Almstrand R, Samuelsson K, Mattsson A, Sörensson F, et al. (2007) Effects of environmental conditions on the nitrifying population dynamics in a pilot wastewater treatment plant. Environ Microbiol 9: 2220-2233. Link: https://goo.gl/9DwVuX
- Bernhard AE, Tucker J, Giblin AE, Stahl DA (2007) Functionally distinct communities of ammonia-oxidizing bacteria along an estuarine salinity gradient. Environ Microbiol 9: 1439-1447. Link: https://goo.gl/XkfgQZ
- de Bie MJM, Speksnijder A, Kowalchuk GA, Schuurma T, Zwart G, et al. (2001) Shifts in the dominant populations of ammonia-oxidizing beta-subclass proteobacteria along the eutrophic Schelde Estuary. Aquat Microb Ecol 23: 225–236. Link: https://goo.gl/fxXFCv
- Rowan AK, Snape JR, Fearnside D, Barer MR, Curtis TP, et al. (2003) Composition and diversity of ammonia-oxidising bacterial communities in wastewater treatment reactors of different design treating identical wastewater. FEMS Microbiol Ecol 43: 195-206. Link: https://goo.gl/CsvrGN
- Wittebolle L, Boon N, Vanparys B, Heylen K, De Vos P, et al. (2005) Failure of the ammonia oxidation process in two pharmaceutical wastewater treatment plants is linked to shifts in the bacterial communities. J Appl Microbiol 99: 997-1006. Link: https://goo.gl/UbhcLL
- Gregory SP, Shields RJ, Fletcher D, Gatland P, Dyson PJ (2010) Bacterial community responses to increasing ammonia concentrations in a recirculating vertical flow saline biofilters. Ecol Engin 36: 1485-1491. Link: https://goo.gl/nhFUJL
- Sudarno U, Bathe S, Winter J, Gallert C (2010) Nitrification in fixed-bed reactors treating saline wastewater. Appl Microbiol Biotechnol 85: 2301-2304. Link: https://goo.gl/K3auDz
- Wittebolle L, Verstraete W, Boon N (2009) The inoculum effect on the ammonia-oxidizing bacterial communities in parallel sequential batch reactors. Wat Res 43: 4149-4158. Link: https://goo.gl/crj6o8
- Sakano Y, Kerkhof L (1998) Assessment of changes in microbial community structure during operation of an ammonia biofilter with molecular tools. Appl Environ Microbiol 64: 4877-4882. Link: https://goo.gl/6ozW3J
- Schramm A, De Beer D, Wagner M, Amann R (1998) Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl Environ Microbiol 64: 3480-3485. Link: https://goo.gl/gVwwNe
- Terada A, Lackner S, Kristensen K, Smets BF (2010) Inoculum effects on community composition and nitritation performance of autotrophic nitrifying biofilm reactors with counter-diffusion geometry. Environ Microbiol 12: 2858-2872. Link: https://goo.gl/ZYdU8E
- Sakami T, Andoh T, Morita T, Yamamoto Y (2012) Phylogenetic diversity of ammonia-oxidizing archaea and bacteria in biofilters of recirculating aquaculture systems. Mar Genomics 7: 27-31. Link: https://goo.gl/9h8bWQ
- Prosser JI (1989) Autotrophic nitrification in bacteria. Adv Microb Physiol 30: 125-181. Link: https://goo.gl/xEXP9e
- Li B, Irvin S, Baker K (2007) The variation of nitrifying bacterial population sizes in a sequencing batch reactor (SBR) treating low, mid, high concentrated synthetic wastewater. J Environ Eng Sci 6: 651-663. Link: https://goo.gl/dm2n5k
- Brown MN, Briones A, Diana J, Raskin L (2013) Ammonia-oxidizing archaea and nitrite-oxidizing nitrospiras in the biofilter of a shrimp recirculating aquaculture system. FEMS Microbiol Ecol83: 17-25. Link: https://goo.gl/tGRzWf
- Sauder LA, Engel K, Stearns JC, Masella AP, Pawliszyn R, et al. (2011) Aquarium nitrification revisited: Thaumarchaeota are the dominant ammonia oxidizers in freshwater aquarium biofilters. PLoS One 6: e23281. Link: https://goo.gl/KrU5TZ
- Ruan YJ, Guo XS, Ye ZY, Liu Y, Zhu SM (2015) Bacterial community analysis of different sections of a biofilter in a full-scale marine recirculating aquaculture system. North Am J Aquaculture 77: 318-326. Link: https://goo.gl/4Cpozf
- Lee DE, Lee J, Kim YM, Myeong JI, Kim KH (2016) Uncultured bacterial diversity in a seawater recirculating aquaculture system revealed by 16S rRNA gene amplicon sequencing. J Microbiol 54: 296-304. Link: https://goo.gl/zSq9vy
- Foesel BU, Gieseke A, Schwermer C, Stief P, Koch L, et al. (2008) Nitrosomonas Nm143-like ammonia oxidizers and Nitrospira marina-like nitrite oxidizers dominate the nitrifier community in a marine aquaculture biofilm. FEMS Microbiol Ecol 63: 192-204. Link: https://goo.gl/7PfRjT
- Keuter S, Kruse M, Lipski A, Spieck E (2011) Relevance of Nitrospira for nitrite oxidation in a marine recirculation aquaculture system and physiological features of a Nitrospira marina-like isolate. Environ Microbiol13: 2536-2547. Link: https://goo.gl/JCC2KU
- Kruse M, Keuter S, Bakker E, Spieck E, Eggers T, et al. (2013) Relevance and Diversity of Nitrospira Populations in Biofilters of Brackish RAS. PLoS ONE 8: e64737. Link: https://goo.gl/C6ZGss
- Alawi M, Off S, Kaya M, Spieck E (2009) Temperature influences the population structure of nitrite-oxidizing bacteria in activated sludge. Environ Microbiol Reports 1: 184–190. Link: https://goo.gl/MynmbJ
- Orso S, Gouy M, Navarro E, Normand P (1994) Molecular Phylogenetic Analysis of Nitrobacter spp. Int J Syst Bact 44: 83-86. Link: https://goo.gl/Zb7h5X
- Alawi M, Lipski A, Sanders T, Pfeiffer EM, Spieck E (2007) Cultivation of a novel cold-adapted nitrite oxidizing betaproteobacterium from the Siberian Arctic. Isme J 1: 256-264. Link: https://goo.gl/6UeiPQ
- Karkman A, Mattila K, Tamminen M, Virta M (2011) Cold temperature decreases bacterial species richness in nitrogen-removing bioreactors treating inorganic mine waters. Biotechnol Bioeng 108: 2876-2883. Link: https://goo.gl/sbenhk
- Lücker S, Schwarz J, Gruber-Dorninger C, Spieck E, Wagner M, et al. (2015) Nitrotoga-like bacteria are previously unrecognized key nitrite oxidizers in full-scale wastewater treatment plants. Isme J 9: 708-720. Link: https://goo.gl/NvWXpL
- Ishii K, Fujitani H, Soh K, Nakagawa T, Takahashi R, et al. (2017) Enrichment and physiological characterization of a cold-adapted nitrite oxidizer Nitrotoga sp. from eelgrass sediments AEM. Link: https://goo.gl/ZLNpfq
- Rurangwa E, Verdegem MCJ (2015) Microorganisms in recirculating aquaculture systems and their management. Rev Aquaculture 7: 117-130. Link: https://goo.gl/szXyuL
- Sayavedra-Soto LA, Arp DJ (2011) Ammonia-oxidizing bacteria: Their biochemistry and molecular biology. In Nitrification. Edited by Ward BB, Arp DJ Klotz MG ASM Press11-37. Link: https://goo.gl/UJU8gs
- Bedard C, Knowles R (1989) Physiology, biochemistry and specific inhibitors of CH4, NH4 and CO Oxidation by methanotrophs and nitrifiers. Microbiol Rev 53: 68–84. Link: https://goo.gl/drwfqs
- Schwermer CU, Ferdelman TG, Stief P, Gieseke A, Rezakhani N, et al. (2010) Effect of nitrate on sulfur transformations in sulfidogenic sludge of a marine aquaculture biofilter. FEMS Microbiol Ecol 72: 476-484. Link: https://goo.gl/K8wJub
- Bejarano-Ortiz DI, Huerta-Ochoa S, Thalasso F, Cuervo-López FM, Texier AC (2015) Kinetic constants for biological ammonium and nitrite oxidation processes under sulfide inhibition. Appl Biochem Biotechnol 177: 1665-1675. Link: https://goo.gl/RP1hpT
- Tsuneda S, Park S, Hayashi H, Jung J, Hirata A (2001) Enhancement of nitrifying biofilm formation using selected EPS produced by heterotrophic bacteria. Wat Sci Technol 43: 197-204. Link: https://goo.gl/VuLcRT
- Summerfelt ST (2003) Ozonation and UV irradiation – an introduction and examples of current applications.Aquacult Eng 28: 21–36. Link: https://goo.gl/iAiczT
- Schröder J, Croot P, Von Dewitz B, Waller,U, Hanel R (2011) Potential and limitations of ozone for the removal of ammonia, nitrite, and yellow substances in marine recirculating aquaculture systems. Aquacult Eng 45: 35-41. Link: https://goo.gl/JCwgwi
- Füssel J, Lam P, Lavik G, Jensen MM, Holtappels M, et al. (2012) Nitrite oxidation in the Namibian oxygen minimum zone. Isme J 6: 1200-1209. Link: https://goo.gl/H6jkpq
- Kindaichi T, Ito T, Okabe S (2004) Ecophysiological interaction between nitrifying bacteria and heterotrophic bacteria in autotrophic nitrifying biofilms as determined by microautoradiography-fluorescence in situ hybridization. Appl Environ Microbiol 70: 1641-1650. Link: https://goo.gl/nVxarS
- Koch H, Luecker S, Albertsen M, Kitzinger K, Herbold C, et al. (2015) Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. PNAS 36: 11371-11376. Link: https://goo.gl/P5NppF
- Biesterfeld S, Figureueroa L (2002) Nitrifying biofilm development with time: activity versus phylogenetic composition. Wat Environ Res 74: 470-479. Link: https://goo.gl/82hbmR
- Van Loosdrecht MCM, Heijnen JJ, Eberl H, Kreft J, Picioreanu C (2002) Mathematical modelling of biofilm structures. Anton Leeuw 81: 245-256. Link: https://goo.gl/S9AyCr
- Lane DJ (1991) 16S/23S rRNA sequencing. In Nucleic acid techniques in bacterial systematics. Edited by Stackebrandt E, Goodfellow M, John Wiley & Sons Chichester, UK, 115-175.
- Muyzer G, Brinkhoff T, Nübel U, Santegoedes H, Schäfer H, Wawer C (1998) Denaturing gradient gel electrophoresis (DGGE) in microbial ecology. In Molecular Microbial Ecology Manual. Edited by Akkermans ADL, van Elsas JD, de Bruijn FJ, Kluwer Academic Publishers, Dordrecht, The Netherlands, 1-27.
- DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89: 5685-5689. Link: https://goo.gl/eQMZZ7
- Mobarry B, Wagner M, Urbain V, Rittmann B, Stahl D (1996) Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl Environ Microbiol 62: 2156-2162. Link: https://goo.gl/knQxUN
- Degrange V, Bardin R (1995) Detection and counting of Nitrobacter populations in soil by PCR. Appl Environ Microbiol 61: 2093-2098. Link: https://goo.gl/cWneRW
- Daims H, PH N, Nielsen JL, Juretschko S, Wagner M (2000) Novel Nitrospira-like bacteria as dominant nitrite-oxidizers in biofilms from wastewater treatment plants: diversity and in situ physiology. Wat Sci Technol 14: 85-90. Link: https://goo.gl/mHNk9M
- Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB (2005) Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci USA102: 14683-14688. Link: https://goo.gl/wExLRo
- Messing J (1983) New M13 vectors for cloning. Methods Enzymol 101: 20-78. Link: https://goo.gl/DBmVjw
- Maixner F, Noguera DR, Anneser B, Stoecker K, Wegl G, et al. (2006) Nitrite concentration influences the population structure of Nitrospira-like bacteria. Environ Microbiol 8: 1487-1495. Link: https://goo.gl/nDQVeU
- Neef A, Amann R, Schlesner H, Schleifer KH (1998) Monitoring a widespread bacterial group: in situ detection of planctomycetes with 16S rRNA-targeted probes. Microbiology 144: 3257-3266. Link: https://goo.gl/Fhy6Ek
- Schmidt I, Zart D, Bock E (2001) Gaseous NO2 as a regulator for ammonia oxidation of Nitrosomonas eutropha. Anton Leeuw79: 311-318. Link: https://goo.gl/Md42do
- Voytek M, Ward BB (1995) Detection of ammonium-oxidizing bacteria of the beta-subclass of the class Proteobacteria in aquatic samples with the PCR. Appl Environ Microbiol 61: 1444-1450. Link: https://goo.gl/7wqRGP

Article Alerts
Subscribe to our articles alerts and stay tuned.
 This work is licensed under a Creative Commons Attribution 4.0 International License.
This work is licensed under a Creative Commons Attribution 4.0 International License.
 Save to Mendeley
Save to Mendeley
